Sullivan Lab: Development & Epigenetics Laboratory
Our research
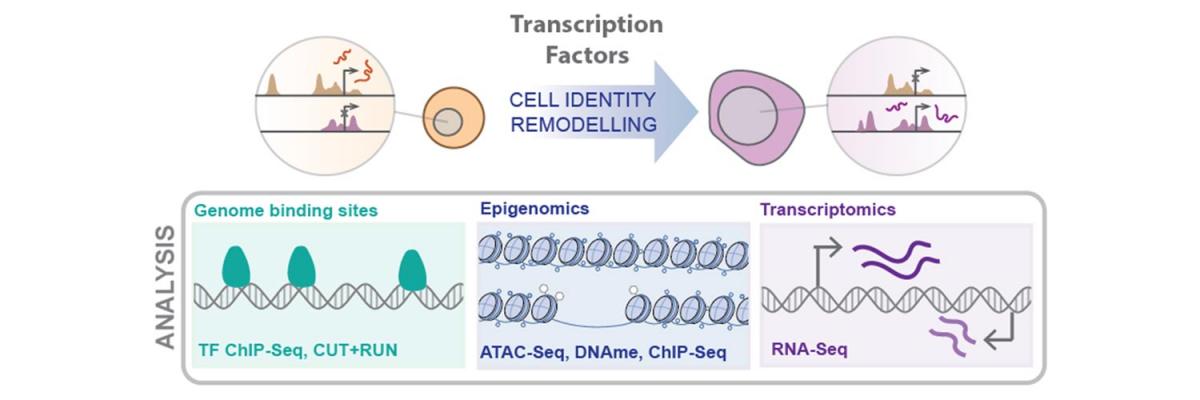
Enhancers are non-coding regions of the genome that are bound by transcription factors in order to control target gene expression. A key paradigm of developmental biology is that cell type-specific gene expression is highly dependent on the availability and selective usage of these enhancer regions, which is controlled through epigenetic modifications.
The profile of active and inactive/silenced enhancers in a cell is therefore a powerful metric for defining cell type, and the selective activation and decommissioning of enhancers through epigenetic remodelling is critical for cells to change identity – for example, in differentiation during development. Conversely, misregulation of enhancers and enhancer remodelling plays a key role in cell transformation in carcinogenesis and can also underly non-genetic drug resistance.
Understanding the molecular biology of these processes and how they translate to cellular properties is therefore important to understand both development and disease.
The Development and Epigenetics Lab seeks to understand the molecular mechanisms controlling activity of enhancers and how this regulation affects the identity, behaviour, and potency of a cell.
Some key questions we are interested in answering are:
- What drives enhancer activation and decommissioning?
- How are these processes misregulated to produce cancerous/pathogenic outcomes?
- How are cellular properties such as lineage potency epigenetically encoded?
To study this key aspect of cellular identity we use complex cell culture-based models, including 3D organoid models, of human development and cancer biology together with advanced multi-omics profiling and precision molecular biology techniques.
We are actively seeking lab members who are excited about this area of research and can work collaboratively, creatively, and with scientific rigor. Students with a keen interest in gene regulation and molecular biology are encouraged to enquire about placement or HDR project opportunities.
Highlighted papers
- Gunne-Braden, A.*, Sullivan, A.*, Gharibi, B., Sheriff, R.S.M., Maity, A., Wang, Y-F., Edwards, A., Jiang, M., Howell, M., Goldstone, R., Wollman, R., East, P. and Santos, S.D.M. (2020) GATA3 mediates a fast, irreversible commitment to BMP4-driven differentiation in human embryonic stem cells. Cell Stem Cell 26(5), 693-706. (*co-first)
- Sullivan, A.E. (2023) Epigenetic control of cell potency and fate determination during mammalian gastrulation. Genes 14(6), 1143
- Sullivan, A.E. & Santos, S.D.M. (2023) The ever-growing world of gastruloids: autogenous models of mammalian embryogenesis. Current Opinion in Genetics & Development 82:102102
- Sullivan, A.E. and Santos, S.D.M. (2020) An Optimized Protocol for ChIP-Seq from Human Embryonic Stem Cell Cultures. STAR Protocols 1(2), Article 100062
