Isbel Lab: Molecular Epigenetics
Our research
How is specificity established in gene regulation and how can we target it?
Epigenetics refers to the forces within our cells that can enable genes to be expressed at the right time, place, and developmental setting. Essentially, it ensures genes that are meant to be active stay active, while genes that are meant to be inactive remain inactive. This intricate process allows multiple tissue and cell types to exist in our bodies that nevertheless all arise from the same genetic material. Our goal is to unravel the molecular mechanisms of how such epigenetic forces drive cellular identity.
Our particular focus centres on transcription factors, which are proteins that ‘read out’ DNA sequence. They are critical for directing the complex gene expression patterns that are established during development. In fact, transcription factors are frequent drivers of cancer when mutated. However, understanding their function in normal or disease settings is challenging because our genomes are not naked - they are wrapped up by histone proteins to form chromatin. An overwhelming amount of evidence suggests that various chromatin states can regulate gene expression, potentially by working hand-in-hand with transcription factors, although the detailed molecular mechanisms remain largely enigmatic.
We are intrigued by interactions between transcription factors and chromatin. By targeting this interface, we aim to identify novel means of altering transcription factor activity to open new avenues of cancer treatment. To achieve this goal, we are using a variety of genomics and proteomics tools in mammalian stem cells, cancer cell lines and animal models. Our research group values inclusivity and curiosity, where we strive for a balance between creativity and scientific rigor that is essential for enabling transformative discoveries.
Lab resources & techniques
- Functional genomics: ChIPseq, ATACseq, RNAseq, Cut and Run, DNA methylomes etc.
- Bioinformatics: Differential expression & binding analysis, motif enrichment analysis.
- Tissue culture: Stem cell and cancer cell line culture.
- In vitro genomics: Selected Engagement on Nucleosome sequencing – SeENseq, DNA affinity purification sequencing – DAPseq.
Projects
We are actively seeking students who bring enthusiasm and great personalities. Inquiries are welcome, both for those with a research question or for those interested in the projects on display below.
-
Dissecting the molecular interplay between transcription factors and chromatin
Project supervisor(s): Dr Luke Isbel
Suitable for: MPhil, PhD
Location of project: Adelaide Health & Medical Sciences Building, North Terrace, AdelaideDescription:
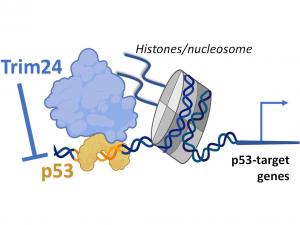
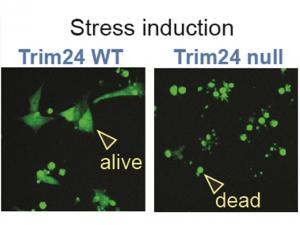
Transcription factors are proteins that bind DNA motifs in regulatory sequences, like enhancers or promoters, to activate expression of nearby genes. At the same time, the genome is embedded in chromatin, where ‘epigenetic’ marks like histone modifications are thought to provide an extra level of gene control. While we suspect there is a remarkable specificity encoded within the interplay between transcription factors and chromatin, the molecular details remain enigmatic. For example, beyond sensing the underlying DNA sequence, do transcription factors also interpret chromatin and can this affect their function?Importantly, transcription factors are often deregulated in cancer, suggesting they can be important therapeutic targets. The tumour suppressor p53 is one such transcription factor and it is frequently silenced in cancer. We recently demonstrated that specific chromatin marks can prevent p53 from activating its target genes, via a mechanism that involves a histone-binding cofactor called Trim24 (Isbel, 2023). The candidate will use this prior work as an experimental platform aimed at addressing the role of chromatin in regulating p53 function, and to link this back to the biology of stress-response in stem cells and colorectal cancer (CRC). They will establish reporters based on target genes of p53 and Trim24 which can be used to screen for additional factors involved in gene regulation, whereupon molecular tools like degron tagging and mutant alleles will be used to investigate their function.
The ultimate goal of this project is to generate a framework for testing both transcription factors and local chromatin marks as a novel therapeutic approach in cancer.
Click image to enlarge.
We will fully support the candidate in learning scientific and technical skills, including a variety of genomic, epigenomic and biochemistry techniques such as CRISPR Cas9 gene-editing, flow cytometry-based genome-wide CRISPR/Cas9 screening, various genomics/epigenetic techniques (i.e. ChIPseq, ATACseq, RNAseq etc.) and sophisticated bioinformatic analysis. This will well-prepare the candidate for a future in science.
Our ideal candidate will be curious about gene regulation and demonstrate creativity in their approach. They should have some experience with basic molecular biology techniques, such as PCR, Western blotting or tissue culture etc. We would strive to recruit a candidate that is excited to shape the success of the group by bringing their own unique experiences and insights.